Select one of your interesting sequences from the database (sequence should be longer than 300 base pair) to do the BLAST search and answer the following questions:
a. What are the different between 6 BLASTs(blastn, blastp, blastx, tblastn, tblastx, PSI-BLAST)?
b. Use your sequence to do 3 out of 6 BLASTs and discuss “What’s the strength and weakness of BLAST you have selected?”
c. Show us the first hit on each BLAST with their identity or/and similarity scores.
d. Summarize the result from 3 BLASTs you select.
a. What are the different between 6 BLASTs(blastn, blastp, blastx, tblastn, tblastx, PSI-BLAST)?
>>Nucleotide-nucleotide BLAST (blastn) :
This program, given a DNA query, returns the most similar DNA sequences from the DNA database that the user specifies. It is used to search very similar DNA sequence.
>>Protein-protein BLAST (blastp):
This program, given a protein query, returns the most similar protein sequences from the protein database that the user specifies. It is used to find out something about the function of the protein.
>>Nucleotide 6-frame translation-protein (blastx):
This program compares the six-frame conceptual translation products of a nucleotide query sequence (both strands) against a protein sequence database. It is used for analysis of the query DNA sequence.
>>Protein-nucleotide 6-frame translation (tblastn):
This program compares a protein query against the all six reading frames of a nucleotide sequence database. It is used to discover new genes encoding simple proteins.
>>Nucleotide 6-frame translation-nucleotide 6-frame translation (tblastx):
This program is the slowest of the BLAST family. It translates the query nucleotide sequence in all six possible frames and compares it against the six-frame translations of a nucleotide sequence database. The purpose of tblastx is to find very distant relationships between nucleotide sequences,protein discovery and ESTs.
>>Position-Specific Iterative BLAST (PSI-BLAST):
This program build profiles and performs databases searches in an iterative fasion. It is used to search protein database using a protein query. It is a very sensitive search strategy to detect weak but biologically significant similarities between sequences. Typically, three to five iterations of PSI-BLAST are sufficient to find most distant homologs at the sequence level.
By including related proteins in the search, PSI-BLAST is much more sensitive in picking up distant evolutionary relationships than a standard protein-protein BLAST.
b. Use your sequence to do 3 out of 6 BLASTs and discuss “What’s the strength and weakness of BLAST you have selected?”
>>blastn, blastx, and tblastx are chosen due to the nature of sequence is a nucleotide sequence.
>>blastn:sequence vs nucleotide database
Strength: possibly more comprehensive. It is mainly used to identify an unknown sequence.
Weakness: lower sequence identity at DNA level.
>>blastx:six-frame translation of your sequence vs protein database
Strength: don’t need to know the right reading frame of your sequence,copes better with (a few) mis-sense errors in your nucleotide sequence.
Weakness: a protein database may not be updated and the coding regions of genomic sequences may not be identified properly.
>>tblastx:six-frame translation of your sequence vs six-frame translation of database.
Strength: tblastx is similar to blastx except a type of database. It uses translated database. It is useful in identifying proteins encoded by single pass read ESTs. It can identify cryptic exons in genomic sequences.
Weakness: slow and a long sequence; for example, a length of 3000 bp is not recommended.
c. Show us the first hit on each BLAST with their identity or/and similarity scores.
>>Selected nucleotide sequence Homo sapiens aldo-keto reductase family 7, member A2 (aflatoxin aldehyde reductase) (AKR7A2), mRNA
. Accession number NM_003689, length: 1377 bp
1.Retrieve the nucleotide sequence of this accession number NM_003689 from GenBank (http://www.ncbi.nlm.nih.gov/). Click on download and choose “FASTA” and save the sequence as a text file.
2.Then, blast the nucleotide sequence with BLAST program available on http://blast.ncbi.nlm.nih.gov/Blast.cgi.
>>Click on “blastn” from this page. Paste the nucleotide sequence in FASTA format.
>>Under “Choose Search Set”, choose “others” checkbox>>nucleotide collection (nr/nt).
>>Under “Program Selection” section, choose “somewhat similar sequence (blastn)” checkbox. Then, click on “BLAST” button. The result page is shown as below.
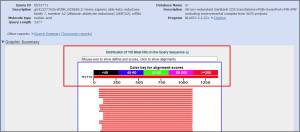
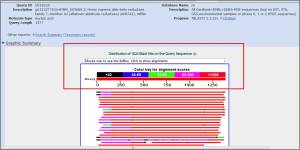
**Graphical overview of pairwise alignments found.The “Distribution of Blast Hits” panel shows how similar the matching sequences are to your input sequence. As the color key indicates, red indicates high similarity, while black indicates relatively low similarity.
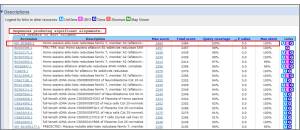
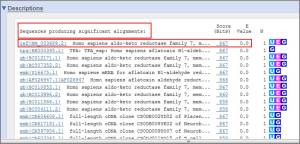
**List of BLAST hits and E-values:The score and E value are listed at the end of each line.The “Sequences producing significant alignments” panel provides you with a list of sequences in the GenBank database that display some similarity to your sequence. They are arranged from most similar to least similar. The “Score” represents a numerical assessment of the similarity two sequences show. The “E value” is a measure of how likely the identified similarity is simply due to chance. The greater the E value is, the less likely it is that the observed similarity is significant.
**BLAST pairwise alignments:The alignments of each “significant” hit to the query. These are shown below in pairwise format.
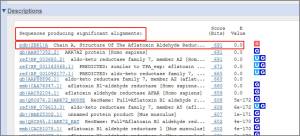
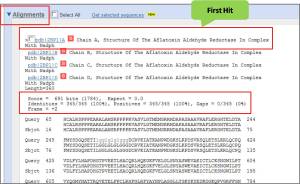
>>For blastn:the first hit is NM_003689 Homo sapiens aldo-keto reductase family 7, member A2 (aflatoxin aldehyde reductase) (AKR7A2), mRNA.Max score = 2484, E-value = 0.0 and Max identity = 100% (1377/1377).
For blastx is done in a similar way to blastn.
Click blastx>>Paste the nucleotide sequence>>database as “nr”>>click on “BLAST” button
>>for blastx:the first hit is 2BP1_A, Chain A, Structure Of The Aflatoxin Aldehyde Reductase In Complex With Nadph, score (Bits) = 691, The identity= 100% (345/345) and E-value = 0.0.
For tblastx is done the same way.
Click tblastx>>paste the nucleotide sequence>>database as “nr/nt”>>click BLAST botton
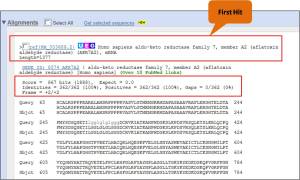
>>for tblastx:the first hit is NM_003689, Homo sapiens aldo-keto reductase family 7, member A2 (aflatoxin aldehyde reductase) (AKR7A2), mRNA, score (Bits) = 867, Identities = 100% (362/362) and E-value = 0.0.
d. Summarize the result from 3 BLASTs you select.
>>The default layout of the NCBI BLAST result is a graphical representation of the hits found, a table of sequence identifiers of the hits together with scoring information, and alignments of the query sequence and the hits.
>>The graphical output gives a quick overview of the query sequence and the resulting hit sequences. The hits are colored according to the obtained alignment scores.The alignments are color coded ranging from black to red as indicated that red shows high similarity, while black shows relatively low similarity.
>>The table view provides more detailed information on each hit, showing the accession number and description field from the sequence file together with BLAST output scores and furthermore acts as a hyperlink to the corresponding sequence in GenBank.
>>In the alignment view one can manually inspect the individual alignments generated by the BLAST algorithm. This is particularly useful for detailed inspection of the sequence hit found(sbjct) and the corresponding alignment. In the alignment view, all scores are described for each alignment, and the start and stop positions for the query and hit sequence are listed. The strand and orientation for query sequence and hits are also found here. The “Score” represents a numerical assessment of the similarity two sequences show. The “E value” is a measure of how likely the identified similarity is simply due to chance.The lower the E-value, the better the hit.
>>Summary of all of blastn, blastx and tblastn show the highest identity 100% matching between query sequence and database sequence. An alignment obtaining an E-value of 0.0 means that identical sequences and there is no chance of occurring by chance alone.