1. What is the name of haploview format to use in this analysis?
HapMap Format
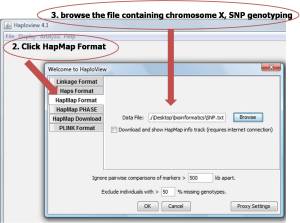
1. Download haploview version 4.1 at http://www.broadinstitute.org/haploview/haploview-downloads
2. Click HapMap format and
3. Browse the file containing the Chromosome X, SNP genotyping (saved data in plain text format (.txt)) and click OK
4. The output data is shown
2. Please show us the marker and individual quality control of the genotype data use in the analysis?
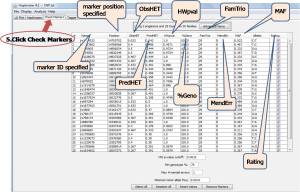
The marker and individual quality control of the genotype data are shown at Check Markers tab
5. Click Check Makers tab
The output data is shown, and the detail of marker is:
– Name: the marker ID specified
– Position: the marker position specified
– ObsHET: the marker’s observed heterozygosity
– PredHET: the marker’s predicted heterozygosity
– HWpval: the Hardy-Weinberg quilibrium p value
-%Geno: the percentage of non-missing genotypes for this marker
– FamTrio: the number of fully genotyped family trios for this marker
– MendErr: the number of observed Mendelian inheritance errors
– MAF: the minor allele frequency for this marker
– Rating is BAD if the marker failed any of the obove tests and blank
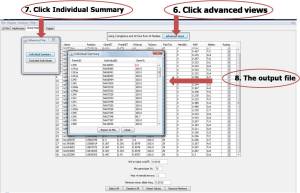
For individual quality control of the genotype data:
6. Click advanced views
7. Click Individual Summary
8. The output data is shown
3. Please show us the LD map then explain what do you get from the LD map?
LD map is shown under LD plot tab
there are 3 regions of triangles grouping red blocks inferring 3 haplotype blocks as shown . The numbers above the map show the marker numbers and names of the alleles.
4. How many haplotype blocks in this region of Chromosome X, then explain how to interpret them?
Haplotypes is shown under Haplotypes tab (1)
Haplotype output displays a block, its markers, the haplotypes and their population frequencies,the crossover percentages to the next block.
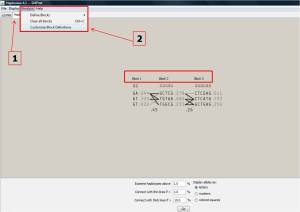
There are 3 haplotype blocks in this region of chromosome X based on 95% confidence, represented as block 1, 2, and 3:
– Block1 contains the marker of 8 and 9
– Block2 contains the marker of 13, 14, 15,16 and 17
– Block3 contains the marker of 24, 25, 26, 27, 28 and 29
Crossing regions show the possibility of recombination between the 2 blocks. The thicker the crossing line,the stronger the recombination.
Besides, the MAF cutoff and confidence bound cutoffs can be edited by selecting customize Block Definitions(2) from Analysis menu
5. Could you find out the tagging SNP in each haplotype block, then explain what the tagging SNPs?
A tag SNP is a representative single nucleotide polymorphism (SNP) in a region of the genome with high linkage disequilibrium
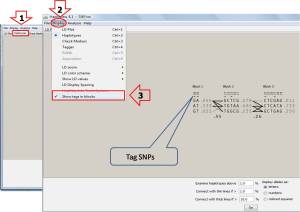
On Haplotypes page (1) , the tag SNPs can be shown by choosing on Display (2) menu and selecting Show tags in blocks (3). Then the tag SNPs are shown by marks under the marker numbers. There are 2 tag SNPs found in each haplotype blocks. Tagging SNPs for haplotype block 1 conatanis GAG and ATT of markers 8 and 9, for haplotype block 2 contains GTT and TTG of markers 13 and 15, and for haplotype block contains CCG and GAG of markers 24 and 27, respectively.
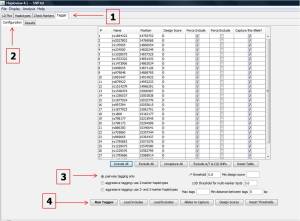
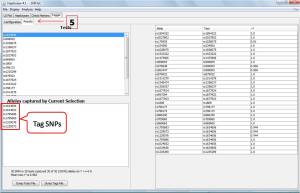
Furthermore, they can be displayed on Tagger page (1) by selecting Configuration tab (2). Then select pairwise tagging only (3) and click on Run Tagger (4) botton and select the Result (5) page. The output data is shown below.
On Alleles captured by Current Selection panel, 6 alleles containing tag SNPs are displayed.



Boy, I am searching the internet and find some elevated communications arrange as forums, blogs. Could you uncover me some suggestion?